FastCCC’s Outputs for Reference-based Analyses
Introduction
Unlike direct CCC analysis on the scRNA-seq dataset, the outputs of reference-based comparison analyses include the following three files:
- The
query_infer_results.tsvfile contains the results of the CCC comparison analysis between the user-provided query data and the reference panel. - The
query_interactions_strength.tsvfile includes the interaction strengths (i.e., communication scores) for each ligand-receptor interaction (columns) across cell-cell interaction pairs (rows) in the query dataset. - The
query_percents_analysis.tsvfile indicates whether the proportion of cells expressing both the ligand and receptor (nonzero expression) exceeds the given threshold in the query dataset.
- The
query_infer_results.tsvfile only includes candidate LRIs where both the ligand and receptor in the corresponding cell types have a nonzero expression proportion exceeding the given threshold (default = 10%).- A significant LRI in the results does not necessarily indicate insignificance in the reference data, and vice versa. The results leverage the accumulated large-scale data to build a reference for a more reliable null distribution. The significance of each LRI in the reference panel (if available) is also reported for comparison.
- The results obtained from the reference comparison and direct anaylsis on the query data may not be identical. However, as demonstrated in our article, the results are stable, and the conclusions are largely consistent. Moreover, for scenarios where the query data has relatively simple cell type composition, using the reference improves accuracy.
- Thus, reference-based analysis is highly effective for scenarios such as examining changes in cell-cell communication between disease and normal states, or when the user-provided query dataset is too small to generate an accurate null distribution.
Outputs of infer_query_workflow
The results of infer_query_workflow are saved to a specified directory, with filenames structured as shown in the figure.

Example of the query_infer_results.tsv file format
For better visualization, we have transposed the dataframe. The row names in the image correspond to the actual column names in the output.
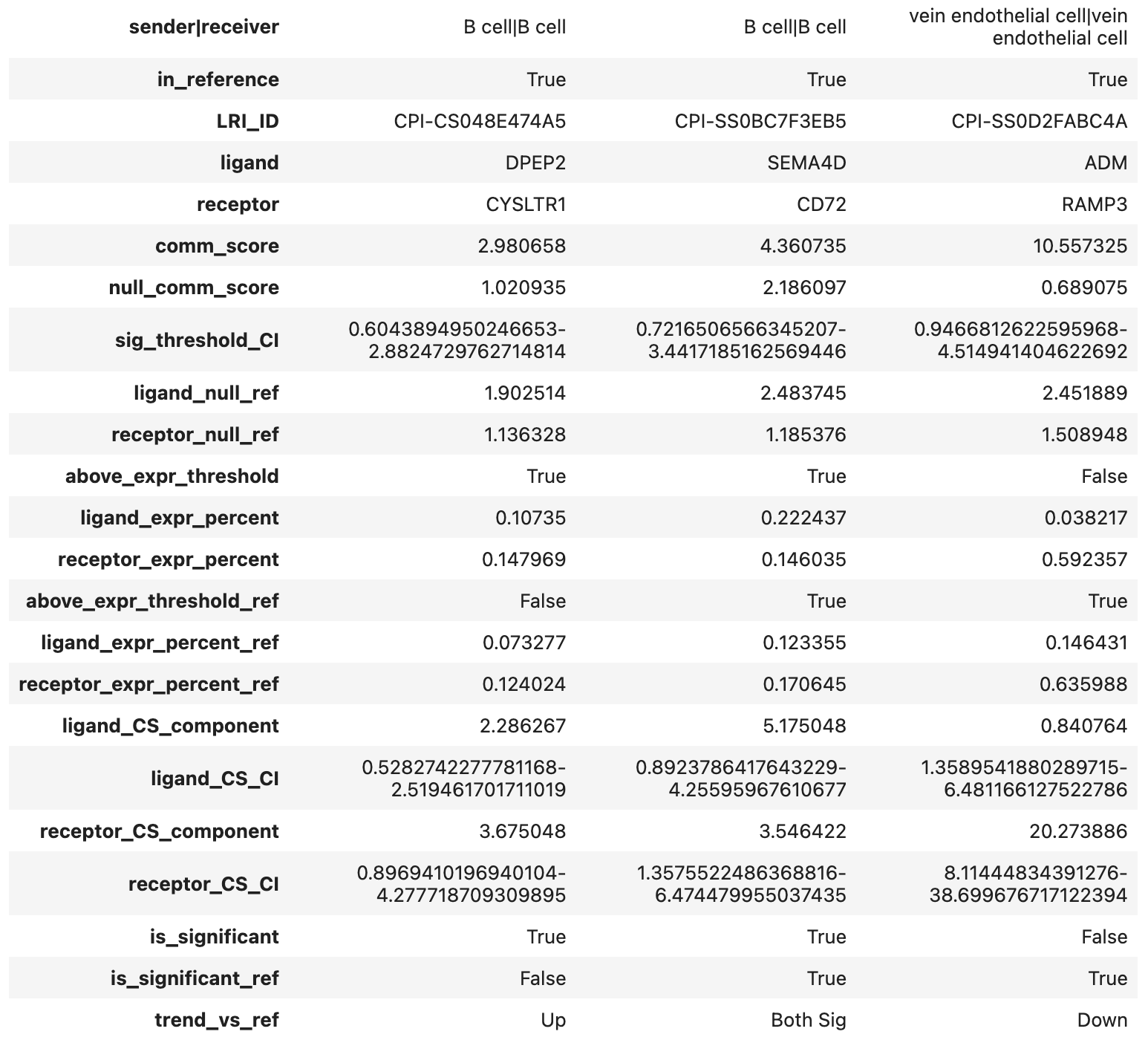
| Column Name | Description |
|---|---|
| sender|receiver | Sender and receiver cell types, separated by |. |
| in_reference | Whether the sender and receiver cell types exist in the reference panel (case-sensitive match required or use celltype_mapping_dict, see here). |
| LRI_ID | Ligand-receptor interaction ID. |
| ligand | Ligand involved in the interaction. |
| receptor | Receptor involved in the interaction. |
| comm_score | Communication score of the ligand-receptor interaction (LRI). |
| null_comm_score | Expected communication score under the null distribution in the query dataset. |
| sig_threshold_CI | 95% confidence interval of the communication score threshold for significance (p = 0.05), inferred from the reference panel. |
| ligand_null_ref | Expected ligand summary score under the null distribution in the reference panel. |
| receptor_null_ref | Expected receptor summary score under the null distribution in the reference panel. |
| above_expr_threshold | Whether both ligand and receptor expression levels exceed the predefined threshold in the query dataset. |
| ligand_expr_percent | Percentage of sender cells expressing the ligand (nonzero expression) in the query dataset. |
| receptor_expr_percent | Percentage of receiver cells expressing the receptor (nonzero expression) in the query dataset. |
| above_expr_threshold_ref | Whether both ligand and receptor expression levels exceed the predefined threshold in the reference dataset. |
| ligand_expr_percent_ref | Percentage of sender cells expressing the ligand in the reference dataset. |
| receptor_expr_percent_ref | Percentage of receiver cells expressing the receptor in the reference dataset. |
| ligand_CS_component | Ligand contribution to the communication score. |
| ligand_CS_CI | 95% CI of ligand expression in the reference dataset, mapped to the query dataset. |
| receptor_CS_component | Receptor contribution to the communication score. |
| receptor_CS_CI | 95% CI of receptor expression in the reference dataset, mapped to the query dataset. |
| is_significant | Whether the ligand-receptor interaction is statistically significant in the query dataset. |
| is_significant_ref | Whether the ligand-receptor interaction is statistically significant in the reference dataset. |
| trend_vs_ref | Change in communication score compared to the reference dataset (e.g., Up for upregulated, Down for downregulated, Both Sig or NS for unchanged). |
Example of the query_interactions_strength.tsv file format
The format of the results is the same as that of the direct CCC analysis. For more details, refer to this link.
Example of the query_percents_analysis.tsv file format
The format of the results is the same as that of the direct CCC analysis. For more details, refer to this link.