FastCCC’s Inputs
Introduction
The functions statistical_analysis_method and Cauchy_combination_of_statistical_analysis_methods require three input datasets:
- Preprocessed scRNA-seq data (log-transformed and stored in .h5ad format).
- Cell type annotations (provided as a tab-separated table or as a column in obsm of the .h5ad file).
- Ligand-receptor interaction (LRI) database (precompiled datasets are available for direct use).
For additional parameters, refer to the Function section. For specific usage examples, refer to the Basic Usage section and Code Snippets.
1. Preprocessed scRNA-seq data
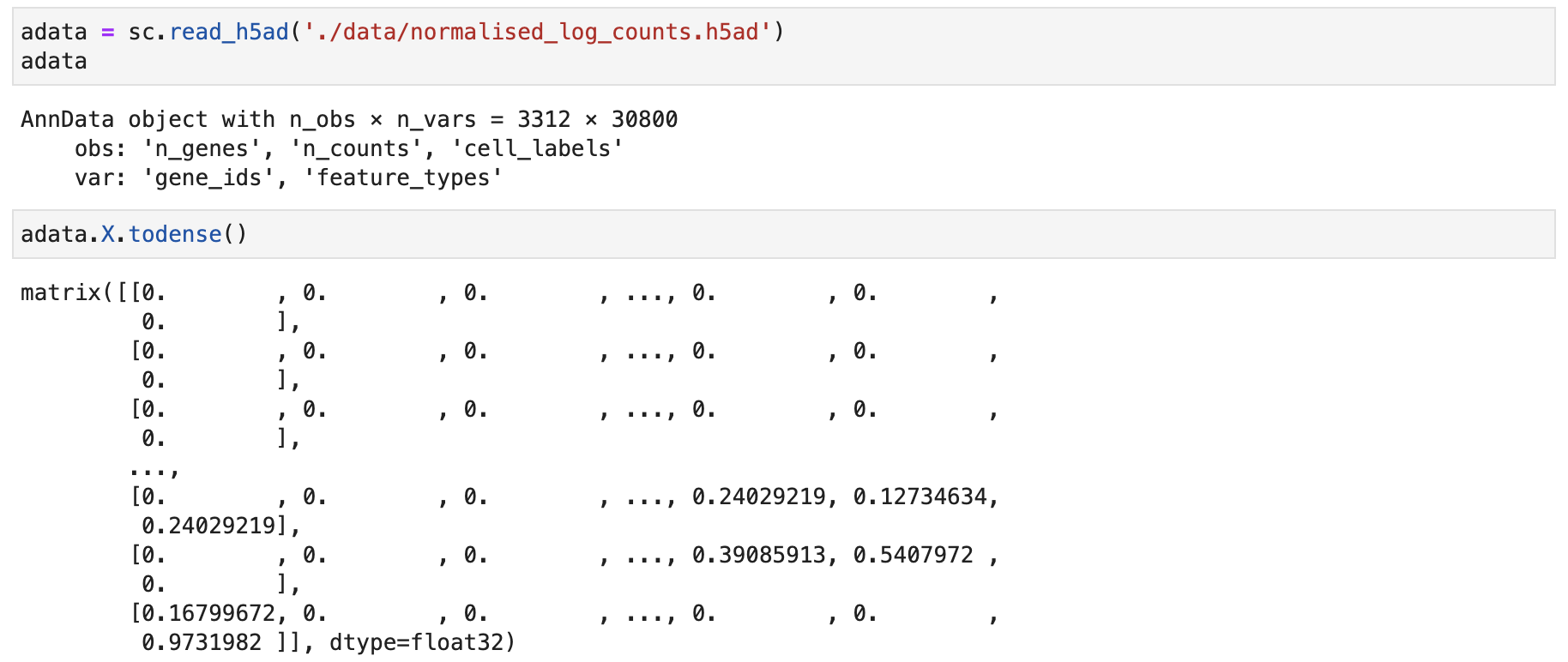
An .h5ad file readable by Scanpy, containing log-transformed (log1p) scRNA-seq data. FastCCC requires this format but does not impose restrictions on other preprocessing steps, such as gene or cell filtering.
Please avoid using HVG (highly variable genes) unless you specifically want to focus on LRI pairs within the HVG set. This allows you to retain a larger number of potential candidate LRI pairs for your analysis.
The internal matrix structure of the scRNA-seq data appears as follows:
2. Cell type annotations
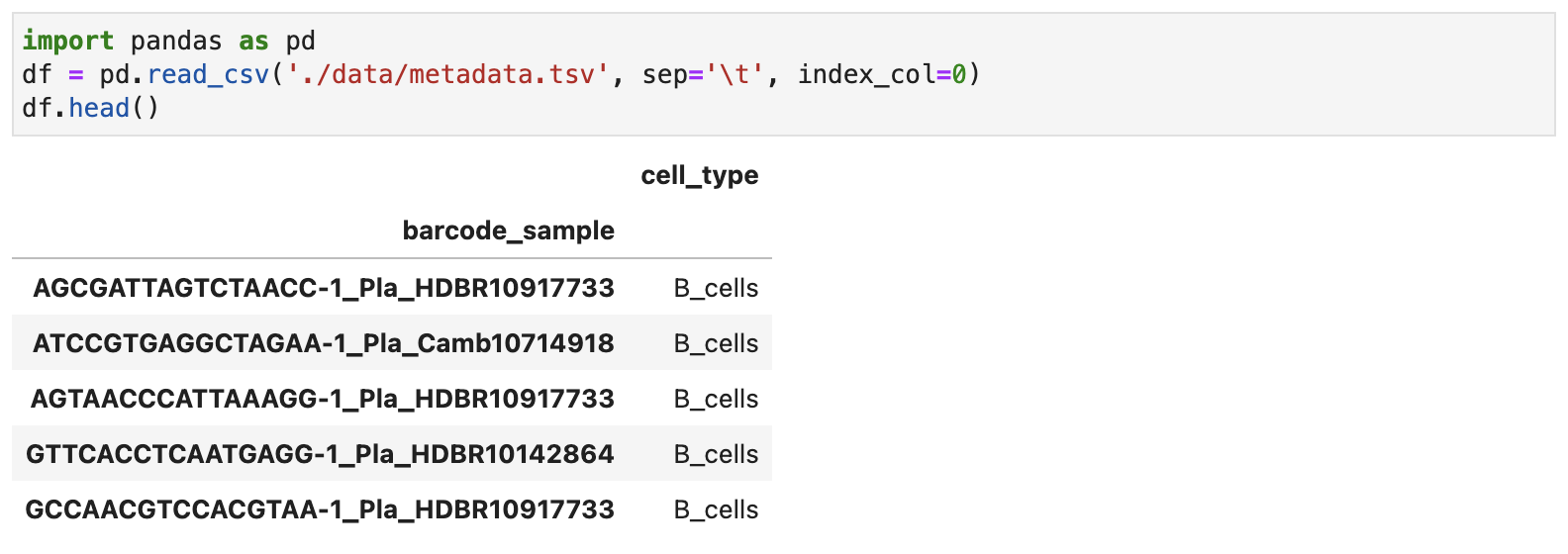
For ease of use, cell type annotations can be provided in two formats. The first option is a TSV (tab-seperated) file, which contains only two columns:
- First column: Cell ID (
barcode_sample), which must match the cell IDs in the .h5ad file. - Second column: Cell type (
cell_type).
The file should be structured as follows:
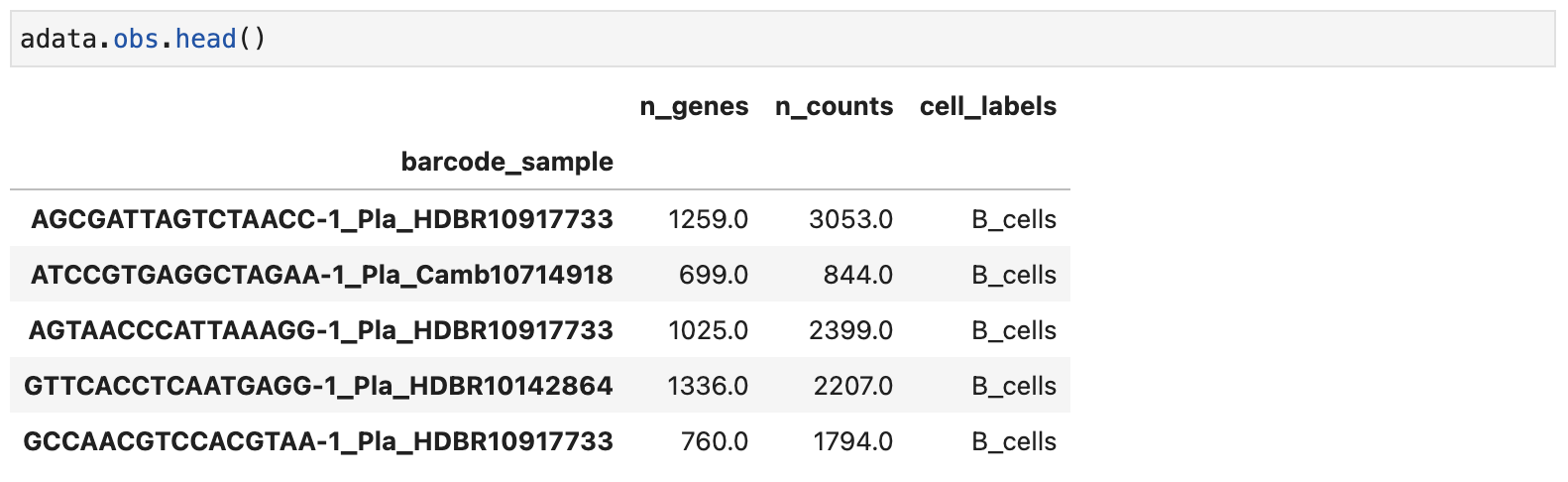
The second option is to use the cell type annotations stored in the .h5ad file’s obs field (if available). For example, as shown in the figure below, the 'cell_labels' column contains the cell type information.
In this case, there is no need to provide a separate TSV file. Instead, simply set
meta_key = 'cell_labels'
3. Ligand-receptor interaction database
We provide preprocessed database on GitHub for users to download directly.
For users who wish to customize their own LRI database, we offer a tool that simplifies the process. You only need to provide a TSV file containing the ligand-receptor interactions, and the tool will automatically generate a properly formatted database file.
To be continued
4. Other required inputs
convert_type
This parameter specifies whether the gene IDs used in your data are HGNC symbols (official gene symbols) or Ensembl IDs.
For example, the 'hgnc_symbol' looks like this:

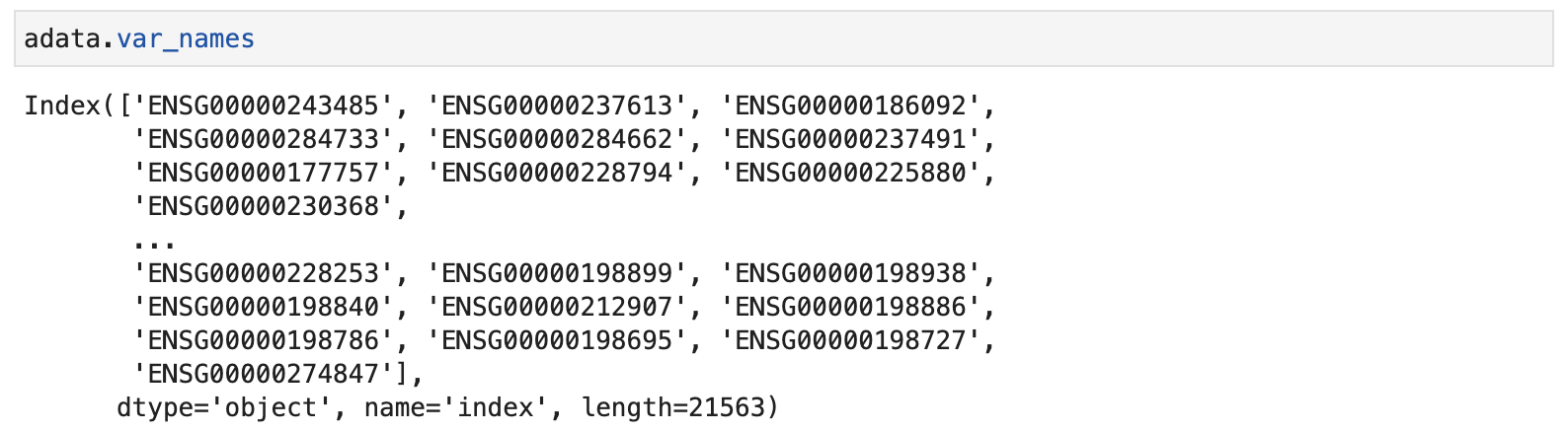
The 'ensembl' looks like this:
For additional parameters, refer to the Function section.